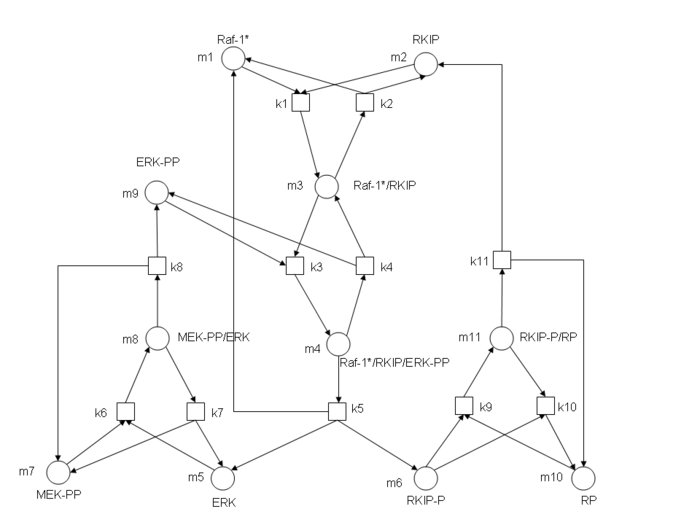
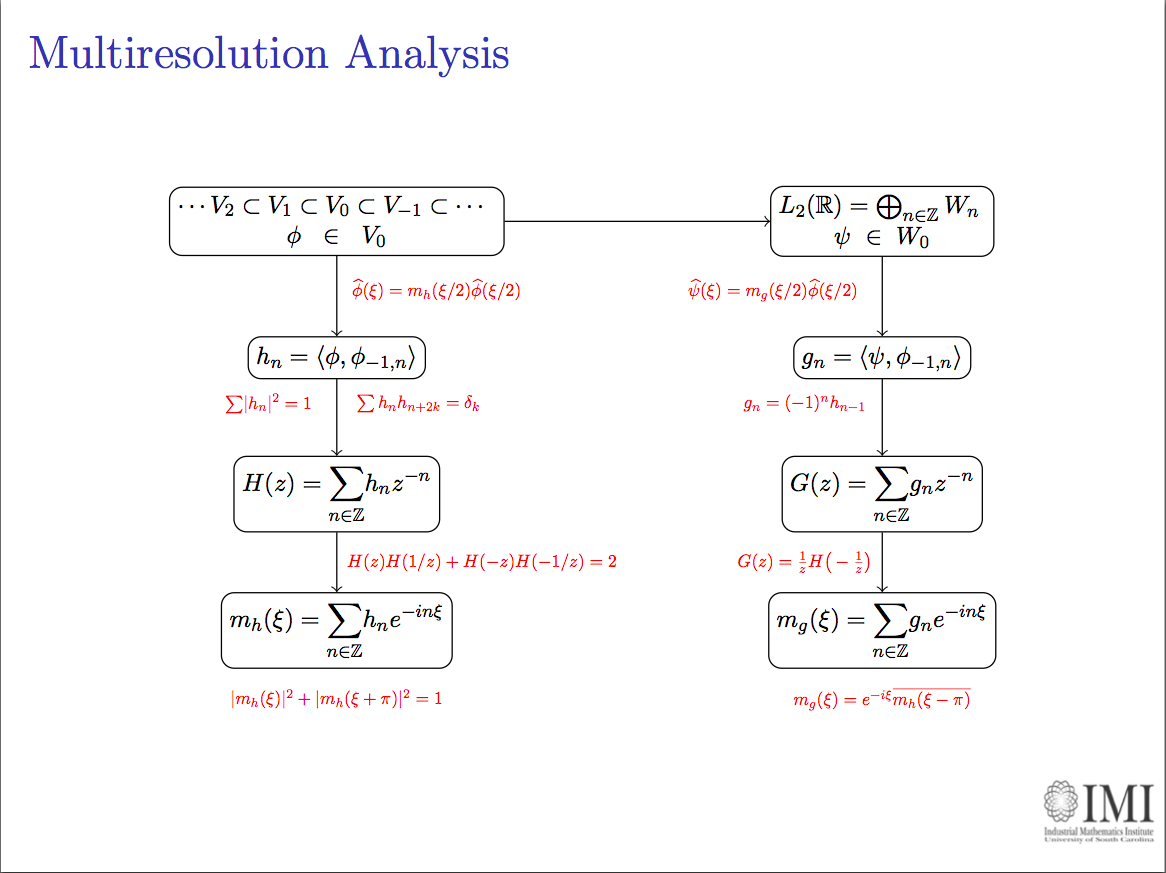
I worked in a project of analysis and simulation of kinetics on networks of enzymatic reactions for a course in BioInformatics. An example: The graph depicted here represents the
RKIP network. Substrates, enzymes and substrate/enzyme complexes are represented by numbered circles, while rate constants are represented by numbered squares.
As a straight-forward application of dynamical systems and numerical solution of systems of differential equations, I wrote a very simple script in
Python which allows a user to input the name of an enzyme, and obtain:
- All networks of chemical reactions in which the input enzyme is present.
- The corresponding (biochemically-correct) systems of algebraic-differential equations expressing the rate of change of the compounds with respect to time.
- Solution to those systems (after user inputs also a set of initial conditions for both the enzymes and substrates)
- Visualization of the process by means of network graphs, plots of amount of chemical compounds with respect to time, etc
Click on the picture above to retrieve a brief report of the project. Find below a concise explanation, and a description of the database
ENZYME, used for the online search of the networks:
Consider the dynamics of a biochemical network of enzymatic reactions: given, from a database, a set of chemical reactions involving related enzymes, each of these take certain amounts of one or several compounds (substrate), and after the reaction, output one or several reactants (notice that the reaction can proceed in either way, depending mainly on the concentration of substrate and reactant). Notice also that both input and output of those reactions might be used by different enzymes in another related reactions.
In this way, if we could “freeze” the reactions at any given time, and measure the concentration of each component being used in this network, we would be able to answer questions of the kind: “Which component decays faster?”, “when will the reactions stop?” (provided the enzymes don’t “die”), “what are the initial concentrations that produce certain phenomena?”, etc.
All these can be answered from a “Dynamical Systems” point of view. Each of the chemical reactions offer a special differential equation involving not only the concentration of the components, but also the concentration of the enzyme and a few more parameters. This differential equations are constrained by the fact that none of the concentrations (=variables) can drop below zero. These are just a small example of the so-called differential-algebraic systems of equations.
Given a network of chemical reactions produced by enzymes, included initial concentrations of components and enzymes, model the dynamical behavior using a differential-algebraic system of equations. The outputs will be, for instance, graphs of the change in the concentration of substrate or reactant A with respect to time.
Of great interest is also the procedure of finding the network itself. For instance, given a certain protein, find all known enzymatic reactions related to the manipulation of this protein. Arrange them in a network, and study its dynamical properties. The first part involves taking information from a database, and a second part involves manipulating that information to obtain the desired network.
ENZYME is a repository of information relative to the nomenclature of enzymes. It is primarily based on the recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (IUBMB) and it describes each type of characterized enzyme for which an EC (Enzyme Commission) number has been provided. It contains the following data:- EC number
- Recommended name
- Alternative names (if any)
- Catalytic activity
- Cofactors (if any)
- Pointers to the SWISS-PROT entry that corresponds to the enzyme (if any)
- Pointers to diseases associated with a deficiency of the enzyme (if any)
The main source for the data in the ENZYME database comes from recommendations of the Nomenclature Committee of the IUBMB.










No comments:
Post a Comment